|
Benjamin R. Jefferys1 |
1 - Imperial College, London |
| Go to 3DPSSM website and submit a protein sequence | Help on using the argumentation system with 3DPSSM | An example which explains the philosophy of argumentation with 3DPSSM | A general description of the theory of argumentation |
This system applies expert knowledge, in the form of an argumentation framework, to results returned by the 3DPSSM Protein Fold Recognition Server. Once a protein sequence has been submitted to that server, the results link to this system. This helps you interpret your search by simplifying and summarising the results, and coming to a conclusion.
This is just one example of a number of possible applications of argumentation, both within bioinformatics and in general. More information on argumentation may be found at the ASPIC website
The 3DPSSM output data used to validate and optimize the argumentation system can be downloaded as a zipped set of XML files here. Documentation is included in the archive and here.
Reference to a paper to cite will appear here
Help using the argumentation system
This section describes a typical session using 3DPSSM with this argumentation system. More information on 3DPSSM can be found on the 3DPSSM help page
1 Submit protein sequence to 3DPSSM
 Enter the protein sequence whose structure you wish to find on the 3DPSSM search page.
Enter the protein sequence whose structure you wish to find on the 3DPSSM search page.
 Once submitted, wait for email confirming completion of your search. This will usually take about half an hour.
Once submitted, wait for email confirming completion of your search. This will usually take about half an hour.
2 View 3DPSSM results
 Click on the link in the result email you receive. This shows you the normal 3DPSSM results. These results may be studied manually, however this is quite hard work and can be simplified using the argumentation system!
Click on the link in the result email you receive. This shows you the normal 3DPSSM results. These results may be studied manually, however this is quite hard work and can be simplified using the argumentation system!
3 View argumentation of 3DPSSM results
 Click on the link for argumentation at the top of the main 3DPSSM results page. There is a Java and non-Java link. The Java version allows you to explore decisions made by the argumentation system. Get Java here.
Click on the link for argumentation at the top of the main 3DPSSM results page. There is a Java and non-Java link. The Java version allows you to explore decisions made by the argumentation system. Get Java here.
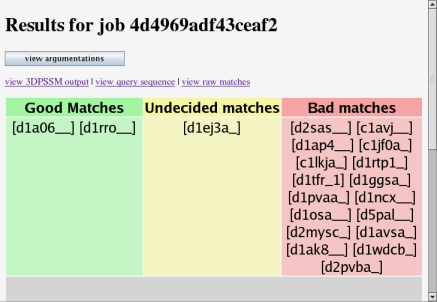 This is the main argumentation results page. The green area contains matches which have been judged as good by the argumentation system. In the yellow area are matches which the system is undecided on. The red area has matches which have been judged as bad. Click on a result to see more information about it.
This is the main argumentation results page. The green area contains matches which have been judged as good by the argumentation system. In the yellow area are matches which the system is undecided on. The red area has matches which have been judged as bad. Click on a result to see more information about it.
 Across the top are links back to the 3DPSSM pages about this search. If you are using the Java version, there is a button to open a new window where the argumentation framework will be displayed when you click on a match.
Across the top are links back to the 3DPSSM pages about this search. If you are using the Java version, there is a button to open a new window where the argumentation framework will be displayed when you click on a match.
4 Explore argumentation for each result
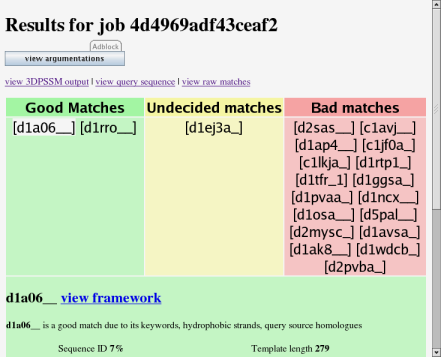 Click on a result name to look at a summary of the argumentation about a search result. The summary appears below the results.
Click on a result name to look at a summary of the argumentation about a search result. The summary appears below the results.
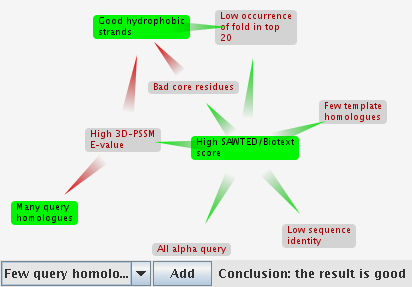 When you select a result, if the Java window is visible, you are shown the argumentation framework. More information about what this diagram means is here. If you don't have Java, you can get it here.
When you select a result, if the Java window is visible, you are shown the argumentation framework. More information about what this diagram means is here. If you don't have Java, you can get it here.
- drag the claims around to change the layout of the diagram
- remove a claim by pointing at it then clicking on the cross that appears to the left
- add a claim by selecting it in the box at the bottom left, and clicking on "Add"
- defeated claims are shown with grey backgrounds
- claims supporting the match are shown in green, and claims against the match are red
- the conclusion of the framework is shown at the bottom right
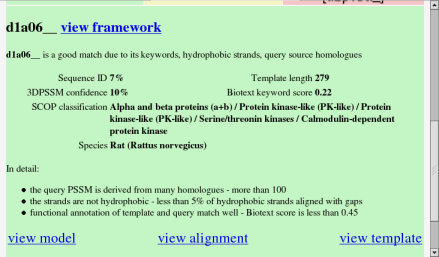 This is the summary of an argumentation for a good result. It shows the following information:
This is the summary of an argumentation for a good result. It shows the following information:
- a link to look at the argumentation framework, without using Java
- a list of features of the match which led the positive conclusion
- some basic information about the match
- undefeated claims about the match which led to the positive conclusion
- links back to 3DPSSM to view the 3D model, alignment and template
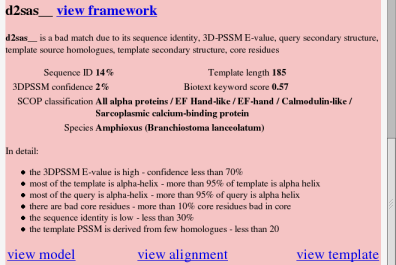 A bad result gives similar information. The features and claims are now those which led to the negative conclusion.
A bad result gives similar information. The features and claims are now those which led to the negative conclusion.
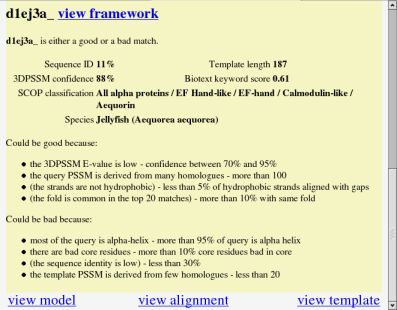 An undecided result gives slightly different information. It gives you all the relevant claims about the match, to help you come to your own conclusion. Some claims are in brackets - these have been defeated by other claims. In this example, "the sequence identity is low" has been defeated by "the 3DPSSM E-value is low" (you can see this by viewing the argumentation framework).
An undecided result gives slightly different information. It gives you all the relevant claims about the match, to help you come to your own conclusion. Some claims are in brackets - these have been defeated by other claims. In this example, "the sequence identity is low" has been defeated by "the 3DPSSM E-value is low" (you can see this by viewing the argumentation framework).
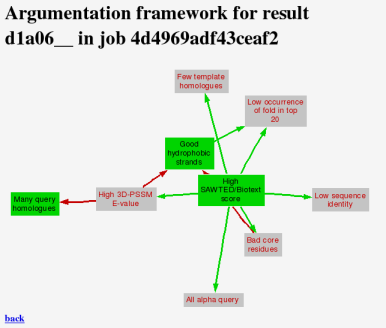 Clicking on "view framework" shows a static view of the argumentation framework, if you cannot use Java. More information about what this diagram means is here.
Clicking on "view framework" shows a static view of the argumentation framework, if you cannot use Java. More information about what this diagram means is here.
- defeated claims are shown with grey backgrounds
- claims supporting the match are shown in green, and claims against the match are red
Worked example - before and after
 This worked example takes the predicted amino acid sequence for a calcium-dependent protein kinase from Pinus taeda (loblolly pine tree - right) and shows the process of finding a putative model for it using 3DPSSM alone, and then applying argumentation to do the same job more easily.
This worked example takes the predicted amino acid sequence for a calcium-dependent protein kinase from Pinus taeda (loblolly pine tree - right) and shows the process of finding a putative model for it using 3DPSSM alone, and then applying argumentation to do the same job more easily.
The protein sequence is as follows:
isggakdlvkrmlehdpkkrlkaadvlshpwmredgeapdkpldstvlvr mkqframnklkkvalkviaeslseeeirglkemfksidadnsgtvtfeel knglarfgskisetevrqlmdavst
Before: 3DPSSM alone
 Go to 3DPSSM submission form and fill in the details as shown, replacing your@email.com with your own email address. Then click "Submit".
Go to 3DPSSM submission form and fill in the details as shown, replacing your@email.com with your own email address. Then click "Submit".
 Eventually you will receive an email like this. Click on the second link to view the results in a web browser.
Eventually you will receive an email like this. Click on the second link to view the results in a web browser.
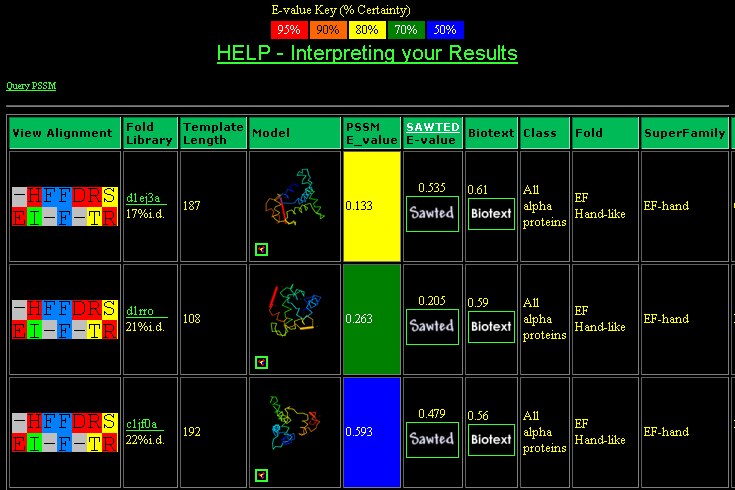 Scroll down and look at the first three results (d1ej3a, d1rro and c1jf0a). Some of the key information is shown here. All three have "EF Hand-like" folds according to SCOP, a fold which appears in most of the searches. The 3DPSSM E-values give 80%, 70% and 50% confidence to the results respectively. The length of the template (the protein found to use as a model for the query sequence) varies by around 85 residues. The percentage identity is quite low for all three. The Sawted and Biotext scores, which reflect how well the functional annotation for the query and template agree, are fairly unimpressive. Further down the page, match d1a06 has a good Biotext score, indicated by the orange background. Click on the link at the left to view the detail of the alignment for d1ej3a.
Scroll down and look at the first three results (d1ej3a, d1rro and c1jf0a). Some of the key information is shown here. All three have "EF Hand-like" folds according to SCOP, a fold which appears in most of the searches. The 3DPSSM E-values give 80%, 70% and 50% confidence to the results respectively. The length of the template (the protein found to use as a model for the query sequence) varies by around 85 residues. The percentage identity is quite low for all three. The Sawted and Biotext scores, which reflect how well the functional annotation for the query and template agree, are fairly unimpressive. Further down the page, match d1a06 has a good Biotext score, indicated by the orange background. Click on the link at the left to view the detail of the alignment for d1ej3a.
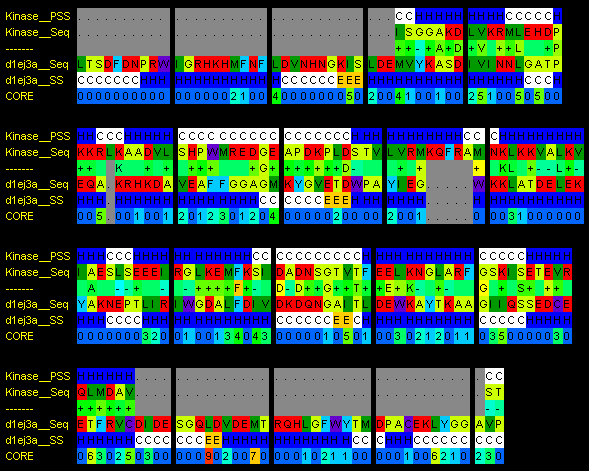 In the alignment for d1ej3a, you can see that there are gaps in the query sequence where core residues in the template structure are supposed to reside, towards the end of the alignment. Coreness is shown in the bottom row, with warmer colours and higher numbers indicating deeper residues (with many contacts). Missing core residues indicate a bad model.
In the alignment for d1ej3a, you can see that there are gaps in the query sequence where core residues in the template structure are supposed to reside, towards the end of the alignment. Coreness is shown in the bottom row, with warmer colours and higher numbers indicating deeper residues (with many contacts). Missing core residues indicate a bad model.
 Go back to the main page and click on "Query PSSM" above the main table. This gives a very long page which shows the multiple sequence alignment used to search for templates by 3DPSSM. The longer it is, the better the search. Scroll to the bottom and you can see there 125 sequences in the PSSM for the query. This is enough to give a very high quality "probe" for the database, giving us some extra confidence in the results.
Go back to the main page and click on "Query PSSM" above the main table. This gives a very long page which shows the multiple sequence alignment used to search for templates by 3DPSSM. The longer it is, the better the search. Scroll to the bottom and you can see there 125 sequences in the PSSM for the query. This is enough to give a very high quality "probe" for the database, giving us some extra confidence in the results.
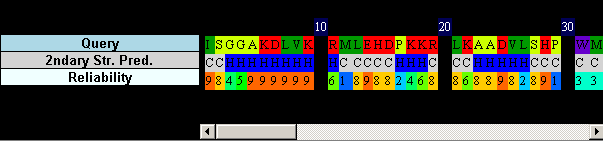 Go back to the main page and look at the bottom of the window, where the query secondary structure is shown. All of the query sequence is predicted as alpha-helix (H) or unstructured coils (C). This makes it easier to align with arbitrary, non-homologous structures, therefore perhaps we should be cautious about all the results.
Go back to the main page and look at the bottom of the window, where the query secondary structure is shown. All of the query sequence is predicted as alpha-helix (H) or unstructured coils (C). This makes it easier to align with arbitrary, non-homologous structures, therefore perhaps we should be cautious about all the results.
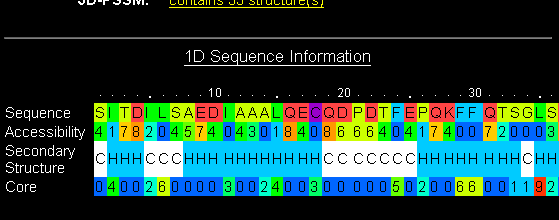 Go back to the main page and click on the link "d1rro" under the column "Fold Library". This shows you the detail about that particular protein template. Look at the 1D Sequence information. Again, most of the structure is alpha-helix or coil, with just a few short strand elements. So perhaps this result is not so reliable.
Go back to the main page and click on the link "d1rro" under the column "Fold Library". This shows you the detail about that particular protein template. Look at the 1D Sequence information. Again, most of the structure is alpha-helix or coil, with just a few short strand elements. So perhaps this result is not so reliable.
With a lot of browsing and a bit of expert knowledge, we've done a very basic analysis of some of the results. In reality we'd have to look more closely at all the results in order to get the most from them. The information we've found is sometimes contradictory - the query sequence is difficult because it is predicted as being all alpha-helix, but is good because it has lots of homologues to use as the probe. These claims affect the confidence we have in all the results, whereas the gaps in core residues and the all-helix template only affect individual results. In all it is a lot of work, and difficult to do fairly and consistently manually. Below we see how argumentation simplifies the process.
After: 3DPSSM and argumentation
 Go back to the main page and click on the link to the "Java" version of the Argumentation system, towards the top of the page. If you do not have Java or have trouble with the Java version, use the "No Java" link. From now on, "Java:" will precede any instructions which only apply if you have Java, and "No Java:" for those when only apply when you don't have Java.
Go back to the main page and click on the link to the "Java" version of the Argumentation system, towards the top of the page. If you do not have Java or have trouble with the Java version, use the "No Java" link. From now on, "Java:" will precede any instructions which only apply if you have Java, and "No Java:" for those when only apply when you don't have Java.
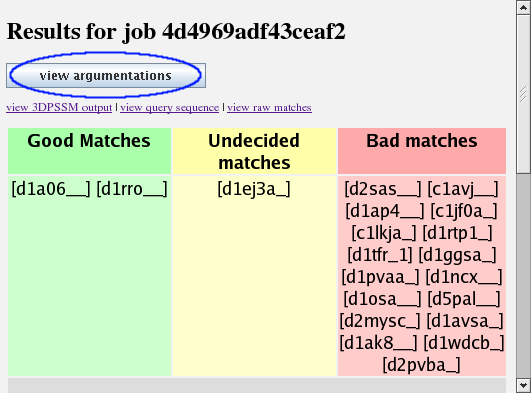 This is the main argumentation page. It gives a very simple summary of what the argumentation system thinks about the results, putting them into three categories: good, bad and undecided.
This is the main argumentation page. It gives a very simple summary of what the argumentation system thinks about the results, putting them into three categories: good, bad and undecided.
Java: Click on the "view argumentations" button to open a new window, and move it so you can see everything.
Now click on [d1a06_] under "Good Matches". It will highlight in white.
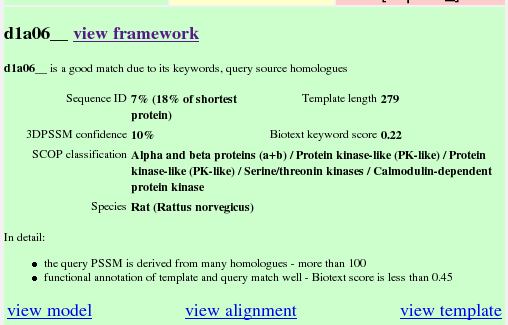 The summary of the argumentation about this result is shown.
The summary of the argumentation about this result is shown.
No Java: Click on the "view framework" link
Recall from the previous section that this result came a long way down the results table, but had a good Biotext score. Also recall that the PSSM for the query was made from many homologues, creating a reliable probe for the database. These two positive features are given in the first sentence: "d1a06__ is a good match due to its keywords, query source homologues". Towards the bottom are some more details about the arguments which mean this is a good match. For example, there were 125 sequences in the query PSSM: this is summarised as "the query PSSM is derived from many homologues", with the justification that there were "more than 100" such homologues.
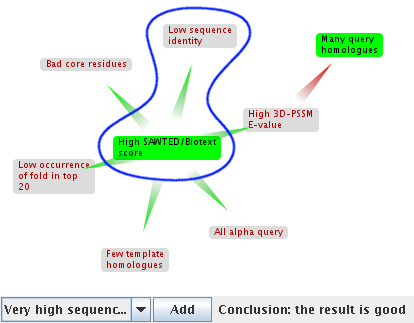 Not all of the claims relevant to a particular match are given in the summary. Some claims are deemed irrelevant as compared to others. For example, d1a06 has a low sequence identity. However this fact is ignored because the good Biotext score is more important. You can see this information in the argumentation framework.
Not all of the claims relevant to a particular match are given in the summary. Some claims are deemed irrelevant as compared to others. For example, d1a06 has a low sequence identity. However this fact is ignored because the good Biotext score is more important. You can see this information in the argumentation framework.
Java: this is in the window you opened earlier. This display is duplicated here.
No Java: this was displayed when you clicked on "view framework". This display is similar to that shown here, which is the Java version.
Circled in blue are these two arguments. The low Biotext score is good for the match, so it is coloured green. The low sequence identity is bad for the match, so it is coloured red. The arrow shows that the Biotext score is more important than the low sequence identity: Biotext attacks low sequence identity, which is shown with a grey background and coloured text to indicate it has been defeated.
 Relationships between the claims can form chains. Here, the claim that there are many query homologues is attacked by a high 3DPSSM E-value: no matter how good the query is, a poor E-value overrides it. However, the good Biotext score in turn attacks the E-value, which is defeated. Its attack on the query homologues is no longer valid, so that claim is not defeated. This process of deciding which claims are defeated is at the core of decision-making with argumentation. Both of the undefeated claims are good for the match, so the overall conclusion of this argumentation is that match d1a06 is good.
Relationships between the claims can form chains. Here, the claim that there are many query homologues is attacked by a high 3DPSSM E-value: no matter how good the query is, a poor E-value overrides it. However, the good Biotext score in turn attacks the E-value, which is defeated. Its attack on the query homologues is no longer valid, so that claim is not defeated. This process of deciding which claims are defeated is at the core of decision-making with argumentation. Both of the undefeated claims are good for the match, so the overall conclusion of this argumentation is that match d1a06 is good.
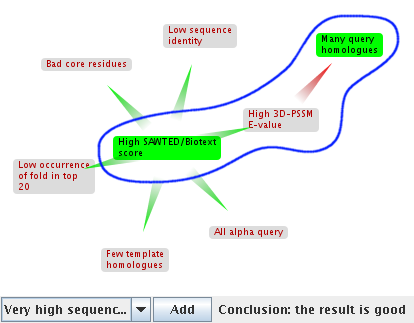
 Java: remove the Biotext claim by pointing at it then clicking on the cross that appears at the top left. It disappears and the argumentation changes radically. The defeated claims change. The query homologues are now defeated, as Biotext isn't there to "rescue" it from the attack by the high 3DPSSM E-value. Since all the undefeated claims are bad for the match, the overall conclusion of the altered argumentation is that match d1a06 is bad.
Java: remove the Biotext claim by pointing at it then clicking on the cross that appears at the top left. It disappears and the argumentation changes radically. The defeated claims change. The query homologues are now defeated, as Biotext isn't there to "rescue" it from the attack by the high 3DPSSM E-value. Since all the undefeated claims are bad for the match, the overall conclusion of the altered argumentation is that match d1a06 is bad.
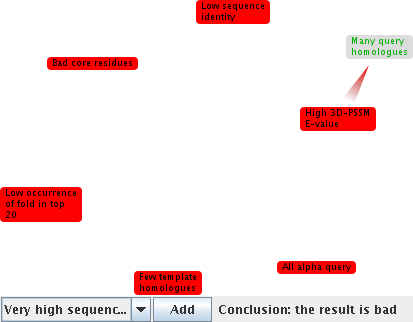
 Move back to the main search results page now, and click on [d1ej3a_] under "Undecided matches". Recall that this was the top match in 3DPSSM, with over 80% confidence. The argumentation says that it might not be such a good match. To find out why...
Move back to the main search results page now, and click on [d1ej3a_] under "Undecided matches". Recall that this was the top match in 3DPSSM, with over 80% confidence. The argumentation says that it might not be such a good match. To find out why...
Java: look at the argumentation in the separate window
No Java: click on "view framework"
This framework shows a mixture of undefeated claims, both good and bad for the match. The system does not try to resolve this problem, instead leaving it to you to interpret the evidence yourself. This diagram may help you.
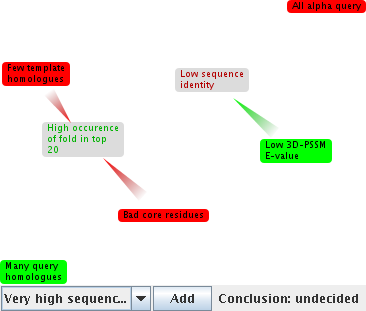
Java: you can experiment with trying to resolve the argumentation by adding or removing claims. For example, you might look at the alignment in 3DPSSM and decide that the core residues aren't so bad. So remove that claim from the diagram. In this case it does not resolve the argumentation. You may also add claims you think apply to the match by selecting them in the box at the bottom left and clicking "Add". Try adding "Very low PSI-Blast E-value": and the argumentation decides that, based on this information, the match is good.
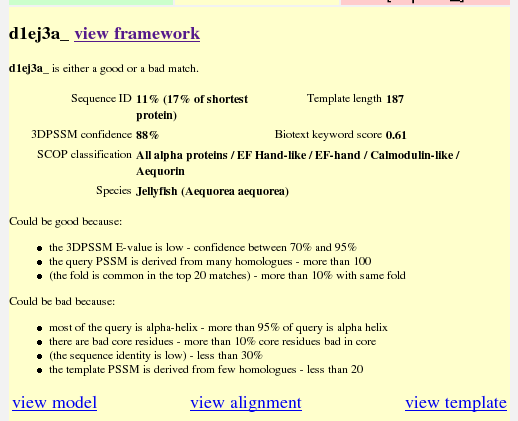 Move back to the main search results page now, and look at the summary of the argumentation for d1ej3a. This tells you all the claims pertinent to the match. The ones in brackets at the bottom are defeated. From this you can draw your own conclusions.
Move back to the main search results page now, and look at the summary of the argumentation for d1ej3a. This tells you all the claims pertinent to the match. The ones in brackets at the bottom are defeated. From this you can draw your own conclusions.
We have used the argumentation system to process and highlight information which previously required a lot of browsing and some expert knowledge. The argumentation system takes the output of 3DPSSM and presents it having been "filtered" to extract and highlight the important information. Matches are sorted into simple categories to highlight likely candidate models. The system allows you to look at the detail of its reasoning process, so you can alter or even dismiss it entirely. Links are provided to return to the 3DPSSM results so you can make your own assessment. It is easy to explore every single result, rather than just a few, so even the "bad" matches may be studied in some detail.
Decision-making with argumentation
This system uses argumentation as a way to come to a final conclusion regarding a particular hypothesis. In our case that hypothesis is "this is a good structural model for the query amino acid sequence". This will be different for other applications. For example, if the method were applied to Google, the hypothesis would be "this is the web page the user is looking for". In medicine it might be "this drug will cure the patient's illness without serious side-effects".
The example below shows how argumentation works by building up a framework, explaining the decisions made at each point. For a more formal study of the process, see Claudette Cayrol, Sylvie Doutre, J�r�me Mengin: On Decision Problems Related to the Preferred Semantics for Argumentation Frameworks. J. Log. Comput. 13(3): 377-403 (2003).
 | 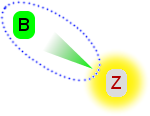 |
| 1. Start with a single claim, Z. Z is coloured red because it is against the hypothesis. There are no other arguments, so it stands uncontested. We conclude the hypothesis is false. | 2. We add a claim B. This is in favour of the hypothesis (green), and it attacks Z (a green arrow). Z has no defence against this attack, so it is defeated (shown in grey) and B is undefeated. We conclude the hypothesis is true. |
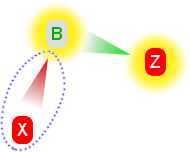 |  |
3. We add a third claim, X, which is against the hypothesis and attacks B (red arrow).
|
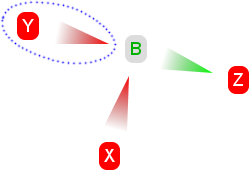
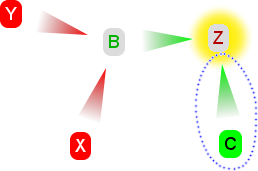
4. A fourth claim is added, called Y. Y is similar to X - it is against the hypothesis and attacks B. This doesn't change our conclusion. X, Y and Z collectively defend themselves against all claims, and all oppose the hypothesis. |
 | 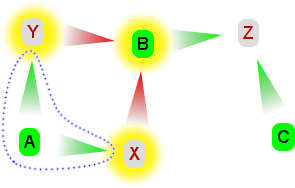 |
| 5. Claim C is added, which is in favour of the hypothesis and attacks Z. X, Y and B are not affected by this addition. However Z is no longer defended against all its attacks: the attack from B is still ignored, but the attack from undefeated C is not. So Z is now defeated. X, Y and C collectively defend themselves against all attacks, however they disagree on the hypothesis. So we cannot come to a conclusion - we are undecided. | 6. Finally we add A. A attacks both X and Y, and is not attacked by anything else. This radically changes the argumentation. X and Y are now defeated. Their attacks on B are now invalid, which is now undefeated. B's attack on Z is now valid, but Z was already defeated anyway so nothing changes there. A, B and C collectively defend themselves against all claims, and they all agree the hypothesis is true, therefore that is our conclusion. |

