1 c3q06B_
100.0
86
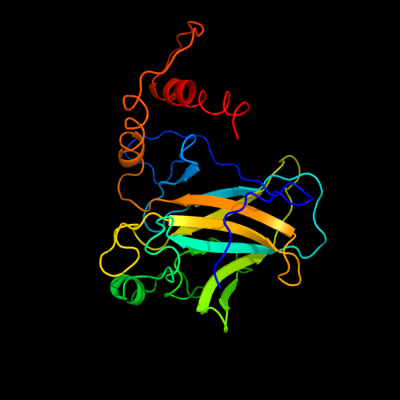
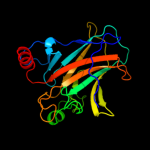
PDB header: cell cycle/dnaChain: B: PDB Molecule: cellular tumor antigen p53;PDBTitle: an induced fit mechanism regulates p53 dna binding kinetics to confer2 sequence specificity
2 c2rmnA_
100.0
51
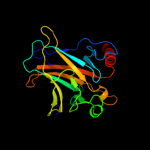
PDB header: cell cycle, antitumor proteinChain: A: PDB Molecule: tumor protein 63;PDBTitle: the solution structure of the p63 dna-binding domain
3 c2j1xA_
100.0
88
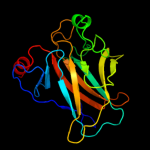
PDB header: nuclear proteinChain: A: PDB Molecule: cellular tumor antigen p53;PDBTitle: human p53 core domain mutant m133l-v203a-y220c-n239y-n268d
4 d2ac0a1
100.0
89
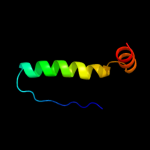
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: p53-like transcription factorsFamily: p53 DNA-binding domain-like5 c2xipA_
100.0
57
PDB header: cell cycleChain: A: PDB Molecule: tumour protein p73;PDBTitle: crystal structure of the dna binding domain of human tp732 refined at 1.8 a resolution
6 d1hu8a_
100.0
99
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: p53-like transcription factorsFamily: p53 DNA-binding domain-like7 c4a9zD_
99.3
31
PDB header: transcriptionChain: D: PDB Molecule: tumor protein 63;PDBTitle: crystal structure of human p63 tetramerization domain
8 c4a9zC_
99.3
31
PDB header: transcriptionChain: C: PDB Molecule: tumor protein 63;PDBTitle: crystal structure of human p63 tetramerization domain
9 c3zy1A_
99.3
30
PDB header: transcriptionChain: A: PDB Molecule: tumor protein 63;PDBTitle: crystal structure of the human p63 tetramerization domain
10 c2wttL_
99.2
30
PDB header: transcriptionChain: L: PDB Molecule: tumor protein p73;PDBTitle: structure of the human p73 tetramerization domain (crystal2 form ii)
11 d3saka_
99.2
86
Fold: p53 tetramerization domainSuperfamily: p53 tetramerization domainFamily: p53 tetramerization domain12 c2wttN_
99.0
32
PDB header: transcriptionChain: N: PDB Molecule: tumor protein p73;PDBTitle: structure of the human p73 tetramerization domain (crystal2 form ii)
13 c2wttH_
99.0
32
PDB header: transcriptionChain: H: PDB Molecule: tumor protein p73;PDBTitle: structure of the human p73 tetramerization domain (crystal2 form ii)
14 c2wqjK_
98.6
43
PDB header: transcriptionChain: K: PDB Molecule: tumor protein p73;PDBTitle: crystal structure of a truncated variant of the human p732 tetramerization domain
15 d1t4wa_
98.6
21
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: p53-like transcription factorsFamily: p53 DNA-binding domain-like16 c2k8fB_
98.6
72
PDB header: transferase/transcriptionChain: B: PDB Molecule: cellular tumor antigen p53;PDBTitle: structural basis for the regulation of p53 function by p300
17 d1hs5a_
98.6
79
Fold: p53 tetramerization domainSuperfamily: p53 tetramerization domainFamily: p53 tetramerization domain18 d1aiea_
98.4
87
Fold: p53 tetramerization domainSuperfamily: p53 tetramerization domainFamily: p53 tetramerization domain19 c2j10B_
98.3
87
PDB header: transcriptionChain: B: PDB Molecule: cellular tumor antigen p53;PDBTitle: p53 tetramerization domain mutant t329f q331k
20 c2j10A_
98.3
87
PDB header: transcriptionChain: A: PDB Molecule: cellular tumor antigen p53;PDBTitle: p53 tetramerization domain mutant t329f q331k
21 c2j10D_
not modelled
98.3
87
PDB header: transcriptionChain: D: PDB Molecule: cellular tumor antigen p53;PDBTitle: p53 tetramerization domain mutant t329f q331k
22 c2j11D_
not modelled
98.3
81
PDB header: transcriptionChain: D: PDB Molecule: cellular tumor antigen p53;PDBTitle: p53 tetramerization domain mutant y327s t329g q331g
23 c2wqjM_
not modelled
98.3
41
PDB header: transcriptionChain: M: PDB Molecule: tumor protein p73;PDBTitle: crystal structure of a truncated variant of the human p732 tetramerization domain
24 d1a1ua_
not modelled
98.2
79
Fold: p53 tetramerization domainSuperfamily: p53 tetramerization domainFamily: p53 tetramerization domain25 c2l14B_
97.7
62
PDB header: protein bindingChain: B: PDB Molecule: cellular tumor antigen p53;PDBTitle: structure of cbp nuclear coactivator binding domain in complex with2 p53 tad
26 c3zy0C_
not modelled
96.9
41
PDB header: transcriptionChain: C: PDB Molecule: tumor protein p63;PDBTitle: crystal structure of a truncated variant of the human p632 tetramerization domain lacking the c-terminal helix
27 c2ly4B_
not modelled
96.9
61
PDB header: nuclear protein/antitumour proteinChain: B: PDB Molecule: cellular tumor antigen p53;PDBTitle: hmgb1-facilitated p53 dna binding occurs via hmg-box/p532 transactivation domain interaction and is regulated by the acidic3 tail
28 c1q2iA_
not modelled
92.1
87
PDB header: antitumor proteinChain: A: PDB Molecule: pnc27;PDBTitle: nmr solution structure of a peptide from the mdm-2 binding2 domain of the p53 protein that is selectively cytotoxic to3 cancer cells
29 c1dt7Y_
91.5
70
PDB header: signaling proteinChain: Y: PDB Molecule: cellular tumor antigen p53;PDBTitle: solution structure of the c-terminal negative regulatory2 domain of p53 in a complex with ca2+-bound s100b(bb)
30 c1dt7X_
91.5
70
PDB header: signaling proteinChain: X: PDB Molecule: cellular tumor antigen p53;PDBTitle: solution structure of the c-terminal negative regulatory2 domain of p53 in a complex with ca2+-bound s100b(bb)
31 c3dacB_
not modelled
67.7
77
PDB header: cell cycleChain: B: PDB Molecule: cellular tumor antigen p53;PDBTitle: structure of the human mdmx protein bound to the p53 tumor2 suppressor transactivation domain
32 c1jspA_
not modelled
63.9
68
PDB header: dna binding proteinChain: A: PDB Molecule: tumor protein p53;PDBTitle: nmr structure of cbp bromodomain in complex with p53 peptide
33 c2xdvA_
not modelled
63.1
32
PDB header: nuclear proteinChain: A: PDB Molecule: myc-induced nuclear antigen;PDBTitle: crystal structure of the catalytic domain of flj14393
34 c3dacP_
not modelled
57.6
83
PDB header: cell cycleChain: P: PDB Molecule: cellular tumor antigen p53;PDBTitle: structure of the human mdmx protein bound to the p53 tumor2 suppressor transactivation domain
35 c4diqA_
not modelled
46.5
40
PDB header: oxidoreductaseChain: A: PDB Molecule: lysine-specific demethylase no66;PDBTitle: crystal structure of human no66
36 c3gycB_
not modelled
25.3
43
PDB header: hydrolaseChain: B: PDB Molecule: putative glycoside hydrolase;PDBTitle: crystal structure of putative glycoside hydrolase (yp_001304622.1)2 from parabacteroides distasonis atcc 8503 at 1.85 a resolution
37 d1vrba1
not modelled
25.1
20
Fold: Double-stranded beta-helixSuperfamily: Clavaminate synthase-likeFamily: Asparaginyl hydroxylase-like38 c4joiA_
not modelled
21.0
21
PDB header: dna binding proteinChain: A: PDB Molecule: cst complex subunit stn1;PDBTitle: crystal structure of the human telomeric stn1-ten1 complex
39 d1iknc_
not modelled
19.7
18
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: NF-kappa-B/REL/DORSAL transcription factors, C-terminal domain40 c3cguB_
not modelled
19.2
44
PDB header: hormone/signaling proteinChain: B: PDB Molecule: protein giant-lens;PDBTitle: crystal structure of unliganded argos
41 c2b3gB_
not modelled
15.6
50
PDB header: replicationChain: B: PDB Molecule: cellular tumor antigen p53;PDBTitle: p53n (fragment 33-60) bound to rpa70n
42 d1ef1c_
not modelled
14.8
21
Fold: Non-globular all-alpha subunits of globular proteinsSuperfamily: Moesin tail domainFamily: Moesin tail domain43 c4gopB_
not modelled
14.7
21
PDB header: dna binding protein/dnaChain: B: PDB Molecule: putative uncharacterized protein;PDBTitle: structure and conformational change of a replication protein a2 heterotrimer bound to ssdna
44 c2pi2A_
not modelled
11.7
11
PDB header: replication, dna binding proteinChain: A: PDB Molecule: replication protein a 32 kda subunit;PDBTitle: full-length replication protein a subunits rpa14 and rpa32
45 d1e7ua3
not modelled
11.1
25
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: Ras-binding domain, RBD46 d1e8ya3
not modelled
11.1
36
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: Ras-binding domain, RBD47 d1w5sa1
not modelled
10.6
25
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Helicase DNA-binding domain48 d2pi2a1
not modelled
10.2
11
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Single strand DNA-binding domain, SSB49 c3kf6A_
not modelled
10.2
13
PDB header: structural proteinChain: A: PDB Molecule: protein stn1;PDBTitle: crystal structure of s. pombe stn1-ten1 complex
50 c3czdA_
not modelled
8.6
42
PDB header: hydrolaseChain: A: PDB Molecule: glutaminase kidney isoform;PDBTitle: crystal structure of human glutaminase in complex with l-glutamate
51 c3ss4C_
not modelled
8.5
42
PDB header: hydrolaseChain: C: PDB Molecule: glutaminase c;PDBTitle: crystal structure of mouse glutaminase c, phosphate-bound form
52 c3ih9A_
not modelled
8.4
29
PDB header: hydrolaseChain: A: PDB Molecule: salt-tolerant glutaminase;PDBTitle: crystal structure analysis of mglu in its tris form
53 c2dfwA_
not modelled
8.2
29
PDB header: hydrolaseChain: A: PDB Molecule: salt-tolerant glutaminase;PDBTitle: crystal structure of a major fragment of the salt-tolerant2 glutaminase from micrococcus luteus k-3
54 c3lsoA_
not modelled
8.1
45
PDB header: membrane proteinChain: A: PDB Molecule: putative membrane anchored protein;PDBTitle: crystal structure of putative membrane anchored protein from2 corynebacterium diphtheriae
55 c1pxeA_
not modelled
8.0
26
PDB header: metal binding proteinChain: A: PDB Molecule: neural zinc finger transcription factor 1;PDBTitle: solution structure of a cchhc domain of neural zinc finger2 factor-1
56 d1regx_
not modelled
8.0
23
Fold: Ferredoxin-likeSuperfamily: Translational regulator protein regAFamily: Translational regulator protein regA57 c2pbyB_
not modelled
7.9
26
PDB header: hydrolaseChain: B: PDB Molecule: glutaminase;PDBTitle: probable glutaminase from geobacillus kaustophilus hta426
58 c1zldA_
not modelled
7.9
54
PDB header: toxinChain: A: PDB Molecule: ptr necrosis toxin;PDBTitle: crystal structure of a rgd-containing host-selective toxin:2 pyrenophora tritici-repentis ptr toxa
59 d1o5wa2
not modelled
7.9
33
Fold: FAD-linked reductases, C-terminal domainSuperfamily: FAD-linked reductases, C-terminal domainFamily: L-aminoacid/polyamine oxidase60 d2je8a3
not modelled
7.9
16
Fold: Immunoglobulin-like beta-sandwichSuperfamily: beta-Galactosidase/glucuronidase domainFamily: beta-Galactosidase/glucuronidase domain61 c3uo9B_
not modelled
7.6
42
PDB header: hydrolase/hydrolase inhibitorChain: B: PDB Molecule: glutaminase kidney isoform, mitochondrial;PDBTitle: crystal structure of human gac in complex with glutamate and bptes
62 d1ko7a2
not modelled
7.4
19
Fold: PEP carboxykinase-likeSuperfamily: PEP carboxykinase-likeFamily: HPr kinase HprK C-terminal domain63 d1ydua1
not modelled
7.2
19
Fold: At5g01610-likeSuperfamily: At5g01610-likeFamily: At5g01610-like64 c1t0pB_
not modelled
6.9
19
PDB header: immune systemChain: B: PDB Molecule: intercellular adhesion molecule-3;PDBTitle: structural basis of icam recognition by integrin alpahlbeta2 revealed2 in the complex structure of binding domains of icam-3 and alphalbeta23 at 1.65 a
65 d1u60a_
not modelled
6.8
29
Fold: beta-lactamase/transpeptidase-likeSuperfamily: beta-lactamase/transpeptidase-likeFamily: Glutaminase66 c2q37A_
not modelled
6.6
30
PDB header: plant protein, lyaseChain: A: PDB Molecule: ohcu decarboxylase;PDBTitle: crystal structure of ohcu decarboxylase in the presence of2 (s)-allantoin
67 d2q37a1
not modelled
6.6
30
Fold: UraD-likeSuperfamily: UraD-LikeFamily: UraD-like68 c1svcP_
not modelled
6.4
15
PDB header: transcription/dnaChain: P: PDB Molecule: protein (nuclear factor kappa-b (nf-kb));PDBTitle: nfkb p50 homodimer bound to dna
69 d1e6vc_
not modelled
6.2
58
Fold: Ferredoxin-likeSuperfamily: Methyl-coenzyme M reductase subunitsFamily: Methyl-coenzyme M reductase gamma chain70 c1ixtA_
not modelled
6.0
80
PDB header: toxinChain: A: PDB Molecule: spasmodic protein tx9a-like protein;PDBTitle: structure of a novel p-superfamily spasmodic conotoxin2 reveals an inhibitory cystine knot motif
71 d1ixta_
not modelled
6.0
80
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: omega toxin-likeFamily: Conotoxin72 d2o8ia1
not modelled
6.0
13
Fold: UraD-likeSuperfamily: UraD-LikeFamily: UraD-like73 d1fx0b1
not modelled
5.7
28
Fold: Left-handed superhelixSuperfamily: C-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: C-terminal domain of alpha and beta subunits of F1 ATP synthase74 d3c7ba1
not modelled
5.5
23
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Ferredoxin domains from multidomain proteins75 d2bosa_
not modelled
5.4
38
Fold: OB-foldSuperfamily: Bacterial enterotoxinsFamily: Bacterial AB5 toxins, B-subunits76 c2wxoA_
not modelled
5.1
27
PDB header: transferaseChain: A: PDB Molecule: phosphatidylinositol-4,5-bisphosphate 3-kinase catalyticPDBTitle: the crystal structure of the murine class ia pi 3-kinase2 p110delta in complex with as5.
77 c3kbyB_
not modelled
5.1
22
PDB header: unknown functionChain: B: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of hypothetical protein from staphylococcus aureus