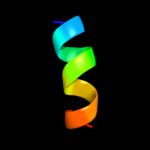| 1 |
|
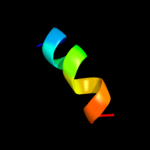
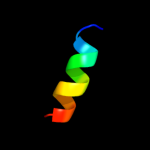
PDB 2axt chain L
Region: 11 - 23
Aligned: 13
Modelled: 13
Confidence: 31.6%
Identity: 38%
PDB header:electron transport
Chain: L: PDB Molecule:photosystem ii reaction center l protein;
PDBTitle: crystal structure of photosystem ii from thermosynechococcus elongatus
Phyre2
| 2 |
|
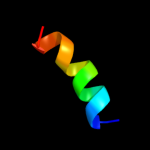
PDB 3kzi chain L
Region: 11 - 23
Aligned: 13
Modelled: 13
Confidence: 31.6%
Identity: 38%
PDB header:electron transport
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: crystal structure of monomeric form of cyanobacterial photosystem ii
Phyre2
| 3 |
|
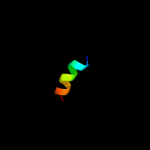
PDB 3a0b chain L
Region: 11 - 23
Aligned: 13
Modelled: 13
Confidence: 31.6%
Identity: 38%
PDB header:electron transport
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: crystal structure of br-substituted photosystem ii complex
Phyre2
| 4 |
|
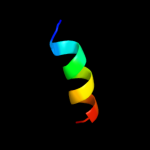
PDB 3prr chain L
Region: 11 - 23
Aligned: 13
Modelled: 13
Confidence: 31.6%
Identity: 38%
PDB header:photosynthesis
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: crystal structure of cyanobacterial photosystem ii in complex with2 terbutryn (part 2 of 2). this file contains second monomer of psii3 dimer
Phyre2
| 5 |
|
PDB 3arc chain L
Region: 11 - 23
Aligned: 13
Modelled: 13
Confidence: 31.6%
Identity: 38%
PDB header:electron transport, photosynthesis
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: crystal structure of oxygen-evolving photosystem ii at 1.9 angstrom2 resolution
Phyre2
| 6 |
|
PDB 1s5l chain L
Region: 11 - 23
Aligned: 13
Modelled: 13
Confidence: 31.6%
Identity: 38%
PDB header:photosynthesis
Chain: L: PDB Molecule:photosystem ii reaction center l protein;
PDBTitle: architecture of the photosynthetic oxygen evolving center
Phyre2
| 7 |
|
PDB 3a0b chain L
Region: 11 - 23
Aligned: 13
Modelled: 13
Confidence: 31.6%
Identity: 38%
PDB header:electron transport
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: crystal structure of br-substituted photosystem ii complex
Phyre2
| 8 |
|
PDB 1s5l chain L
Region: 11 - 23
Aligned: 13
Modelled: 13
Confidence: 31.6%
Identity: 38%
PDB header:photosynthesis
Chain: L: PDB Molecule:photosystem ii reaction center l protein;
PDBTitle: architecture of the photosynthetic oxygen evolving center
Phyre2
| 9 |
|
PDB 2axt chain L
Region: 11 - 23
Aligned: 13
Modelled: 13
Confidence: 31.6%
Identity: 38%
PDB header:electron transport
Chain: L: PDB Molecule:photosystem ii reaction center l protein;
PDBTitle: crystal structure of photosystem ii from thermosynechococcus elongatus
Phyre2
| 10 |
|
PDB 3bz1 chain L
Region: 11 - 23
Aligned: 13
Modelled: 13
Confidence: 31.6%
Identity: 38%
PDB header:electron transport
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: crystal structure of cyanobacterial photosystem ii (part 12 of 2). this file contains first monomer of psii dimer
Phyre2
| 11 |
|
PDB 3prq chain L
Region: 11 - 23
Aligned: 13
Modelled: 13
Confidence: 31.6%
Identity: 38%
PDB header:photosynthesis
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: crystal structure of cyanobacterial photosystem ii in complex with2 terbutryn (part 1 of 2). this file contains first monomer of psii3 dimer
Phyre2
| 12 |
|
PDB 3bz2 chain L
Region: 11 - 23
Aligned: 13
Modelled: 13
Confidence: 31.6%
Identity: 38%
PDB header:electron transport
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: crystal structure of cyanobacterial photosystem ii (part 22 of 2). this file contains second monomer of psii dimer
Phyre2
| 13 |
|
PDB 2axt chain L domain 1
Region: 11 - 23
Aligned: 13
Modelled: 13
Confidence: 31.6%
Identity: 38%
Fold: Single transmembrane helix
Superfamily: Photosystem II reaction center protein L, PsbL
Family: PsbL-like
Phyre2
| 14 |
|
PDB 3bz1 chain Y
Region: 3 - 16
Aligned: 14
Modelled: 14
Confidence: 23.1%
Identity: 57%
PDB header:electron transport
Chain: Y: PDB Molecule:photosystem ii protein y;
PDBTitle: crystal structure of cyanobacterial photosystem ii (part 12 of 2). this file contains first monomer of psii dimer
Phyre2
| 15 |
|
PDB 3a0h chain Y
Region: 3 - 16
Aligned: 14
Modelled: 14
Confidence: 23.1%
Identity: 57%
PDB header:electron transport
Chain: Y: PDB Molecule:photosystem ii reaction center protein ycf12;
PDBTitle: crystal structure of i-substituted photosystem ii complex
Phyre2
| 16 |
|
PDB 3a0h chain Y
Region: 3 - 16
Aligned: 14
Modelled: 14
Confidence: 23.1%
Identity: 57%
PDB header:electron transport
Chain: Y: PDB Molecule:photosystem ii reaction center protein ycf12;
PDBTitle: crystal structure of i-substituted photosystem ii complex
Phyre2
| 17 |
|
PDB 3arc chain Y
Region: 3 - 16
Aligned: 14
Modelled: 14
Confidence: 22.1%
Identity: 57%
PDB header:electron transport, photosynthesis
Chain: Y: PDB Molecule:protein ycf12;
PDBTitle: crystal structure of oxygen-evolving photosystem ii at 1.9 angstrom2 resolution
Phyre2
| 18 |
|
PDB 3arc chain Y
Region: 3 - 16
Aligned: 14
Modelled: 14
Confidence: 22.1%
Identity: 57%
PDB header:electron transport, photosynthesis
Chain: Y: PDB Molecule:protein ycf12;
PDBTitle: crystal structure of oxygen-evolving photosystem ii at 1.9 angstrom2 resolution
Phyre2
| 19 |
|
PDB 3arc chain L
Region: 9 - 23
Aligned: 15
Modelled: 15
Confidence: 22.0%
Identity: 33%
PDB header:electron transport, photosynthesis
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: crystal structure of oxygen-evolving photosystem ii at 1.9 angstrom2 resolution
Phyre2
| 20 |
|
PDB 3a0h chain L
Region: 11 - 23
Aligned: 13
Modelled: 13
Confidence: 21.8%
Identity: 38%
PDB header:electron transport
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: crystal structure of i-substituted photosystem ii complex
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|