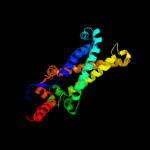
| 1 | d1kqfc_ |
|
 |
100.0 |
100 |
Fold:Heme-binding four-helical bundle
Superfamily:Transmembrane di-heme cytochromes
Family:Formate dehydrogenase N, cytochrome (gamma) subunit |
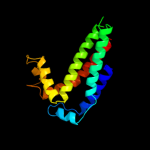
| 2 | d3cx5c2 |
|
 |
97.8 |
12 |
Fold:Heme-binding four-helical bundle
Superfamily:Transmembrane di-heme cytochromes
Family:Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase) |
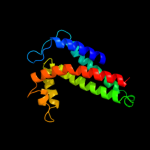
| 3 | c2qjkM_ |
|
 |
97.7 |
14 |
PDB header:electron transport
Chain: M: PDB Molecule:cytochrome b;
PDBTitle: crystal structure analysis of mutant rhodobacter2 sphaeroides bc1 with stigmatellin and antimycin
|
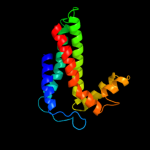
| 4 | d1q90b_ |
|
 |
97.6 |
11 |
Fold:Heme-binding four-helical bundle
Superfamily:Transmembrane di-heme cytochromes
Family:Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase) |
| 5 | d2e74a1 |
|
 |
97.6 |
14 |
Fold:Heme-binding four-helical bundle
Superfamily:Transmembrane di-heme cytochromes
Family:Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase) |
| 6 | c3cx5N_ |
|
 |
97.5 |
13 |
PDB header:oxidoreductase
Chain: N: PDB Molecule:cytochrome b;
PDBTitle: structure of complex iii with bound cytochrome c in reduced2 state and definition of a minimal core interface for3 electron transfer.
|
| 7 | d1ppjc2 |
|
 |
97.4 |
16 |
Fold:Heme-binding four-helical bundle
Superfamily:Transmembrane di-heme cytochromes
Family:Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase) |
| 8 | d1bccc3 |
|
 |
97.4 |
15 |
Fold:Heme-binding four-helical bundle
Superfamily:Transmembrane di-heme cytochromes
Family:Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase) |
| 9 | c3cwbC_ |
|
 |
97.3 |
13 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:cytochrome b;
PDBTitle: chicken cytochrome bc1 complex inhibited by an iodinated analogue of2 the polyketide crocacin-d
|
| 10 | d1y5ic1 |
|
 |
38.1 |
15 |
Fold:Heme-binding four-helical bundle
Superfamily:Respiratory nitrate reductase 1 gamma chain
Family:Respiratory nitrate reductase 1 gamma chain |
| 11 | c2kygC_ |
|
 |
16.4 |
23 |
PDB header:protein binding
Chain: C: PDB Molecule:protein cbfa2t1;
PDBTitle: structure of the aml1-eto nervy domain - pka(riia) complex and its2 contribution to aml1-eto activity
|
| 12 | c2levA_ |
|
 |
13.3 |
47 |
PDB header:transcription regulator/dna
Chain: A: PDB Molecule:ler;
PDBTitle: structure of the dna complex of the c-terminal domain of ler
|
| 13 | d1ecfa1 |
|
 |
11.1 |
9 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 14 | c1fcuA_ |
|
 |
8.2 |
29 |
PDB header:hydrolase
Chain: A: PDB Molecule:hyaluronoglucosaminidase;
PDBTitle: crystal structure (trigonal) of bee venom hyaluronidase
|
| 15 | c1ecjB_ |
|
 |
7.8 |
9 |
PDB header:transferase
Chain: B: PDB Molecule:glutamine phosphoribosylpyrophosphate
PDBTitle: escherichia coli glutamine phosphoribosylpyrophosphate2 (prpp) amidotransferase complexed with 2 amp per tetramer
|
| 16 | c2xzn2_ |
|
 |
7.7 |
10 |
PDB header:ribosome
Chain: 2: PDB Molecule:40s ribosomal protein s8;
PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 2
|
| 17 | d1fcqa_ |
|
 |
7.6 |
29 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Bee venom hyaluronidase |
| 18 | c2pe4A_ |
|
 |
6.3 |
29 |
PDB header:hydrolase
Chain: A: PDB Molecule:hyaluronidase-1;
PDBTitle: structure of human hyaluronidase 1, a hyaluronan hydrolyzing enzyme2 involved in tumor growth and angiogenesis
|
| 19 | c3nrtC_ |
|
 |
6.0 |
21 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:putative ryanodine receptor;
PDBTitle: the crystal strucutre of putative ryanodine receptor from bacteroides2 thetaiotaomicron vpi-5482
|
| 20 | d1m56d_ |
|
 |
5.8 |
12 |
Fold:Single transmembrane helix
Superfamily:Bacterial aa3 type cytochrome c oxidase subunit IV
Family:Bacterial aa3 type cytochrome c oxidase subunit IV |
| 21 | c2qfaC_ |
|
not modelled |
5.7 |
38 |
PDB header:cell cycle/cell cycle/cell cycle
Chain: C: PDB Molecule:inner centromere protein;
PDBTitle: crystal structure of a survivin-borealin-incenp core complex
|
| 22 | c1v0dA_ |
|
not modelled |
5.4 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:dna fragmentation factor 40 kda subunit;
PDBTitle: crystal structure of caspase-activated dnase (cad)
|
| 23 | d1v0da_ |
|
not modelled |
5.4 |
21 |
Fold:His-Me finger endonucleases
Superfamily:His-Me finger endonucleases
Family:Caspase-activated DNase, CAD (DffB, DFF40) |
| 24 | d2ct5a1 |
|
not modelled |
5.4 |
26 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:BED zinc finger |
| 25 | c1rrzA_ |
|
not modelled |
5.3 |
20 |
PDB header:structural genomics,biosynthetic protein
Chain: A: PDB Molecule:glycogen synthesis protein glgs;
PDBTitle: solution structure of glgs protein from e. coli
|
| 26 | d1rrza_ |
|
not modelled |
5.3 |
20 |
Fold:Spectrin repeat-like
Superfamily:Glycogen synthesis protein GlgS
Family:Glycogen synthesis protein GlgS |

