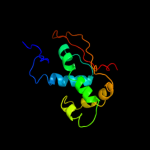
| 1 | d2qamn1 |
|
 |
100.0 |
100 |
Fold:Prokaryotic ribosomal protein L17
Superfamily:Prokaryotic ribosomal protein L17
Family:Prokaryotic ribosomal protein L17 |
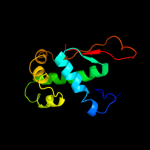
| 2 | c3bboP_ |
|
 |
100.0 |
45 |
PDB header:ribosome
Chain: P: PDB Molecule:ribosomal protein l17;
PDBTitle: homology model for the spinach chloroplast 50s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome
|
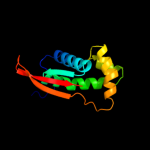
| 3 | d2zjrk1 |
|
 |
100.0 |
52 |
Fold:Prokaryotic ribosomal protein L17
Superfamily:Prokaryotic ribosomal protein L17
Family:Prokaryotic ribosomal protein L17 |
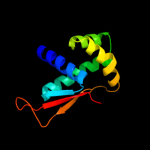
| 4 | d1gd8a_ |
|
 |
100.0 |
53 |
Fold:Prokaryotic ribosomal protein L17
Superfamily:Prokaryotic ribosomal protein L17
Family:Prokaryotic ribosomal protein L17 |
| 5 | d2cqma1 |
|
 |
100.0 |
34 |
Fold:Prokaryotic ribosomal protein L17
Superfamily:Prokaryotic ribosomal protein L17
Family:Prokaryotic ribosomal protein L17 |
| 6 | c3h0dB_ |
|
 |
19.3 |
50 |
PDB header:transcription/dna
Chain: B: PDB Molecule:ctsr;
PDBTitle: crystal structure of ctsr in complex with a 26bp dna duplex
|
| 7 | d1xd7a_ |
|
 |
15.4 |
14 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Transcriptional regulator Rrf2 |
| 8 | d1o65a_ |
|
 |
15.0 |
25 |
Fold:PK beta-barrel domain-like
Superfamily:PK beta-barrel domain-like
Family:MOSC (MOCO sulphurase C-terminal) domain |
| 9 | c2op8A_ |
|
 |
14.1 |
5 |
PDB header:isomerase
Chain: A: PDB Molecule:probable tautomerase ywhb;
PDBTitle: crystal structure of ywhb- homologue of 4-oxalocrotonate tautomerase
|
| 10 | c2z99A_ |
|
 |
13.5 |
26 |
PDB header:cell cycle
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of scpb from mycobacterium tuberculosis
|
| 11 | c2ic6B_ |
|
 |
13.3 |
23 |
PDB header:viral protein
Chain: B: PDB Molecule:nucleocapsid protein;
PDBTitle: the coiled-coil domain (residues 1-75) structure of the sin2 nombre virus nucleocapsid protein
|
| 12 | c1x4qA_ |
|
 |
10.9 |
26 |
PDB header:rna binding protein
Chain: A: PDB Molecule:u4/u6 small nuclear ribonucleoprotein prp3;
PDBTitle: solution structure of pwi domain in u4/u6 small nuclear2 ribonucleoprotein prp3(hprp3)
|
| 13 | d1v66a_ |
|
 |
10.8 |
26 |
Fold:LEM/SAP HeH motif
Superfamily:SAP domain
Family:SAP domain |
| 14 | d1otfa_ |
|
 |
9.9 |
10 |
Fold:Tautomerase/MIF
Superfamily:Tautomerase/MIF
Family:4-oxalocrotonate tautomerase-like |
| 15 | d1bjpa_ |
|
 |
8.9 |
14 |
Fold:Tautomerase/MIF
Superfamily:Tautomerase/MIF
Family:4-oxalocrotonate tautomerase-like |
| 16 | c3ry0A_ |
|
 |
8.7 |
10 |
PDB header:isomerase
Chain: A: PDB Molecule:putative tautomerase;
PDBTitle: crystal structure of tomn, a 4-oxalocrotonate tautomerase homologue in2 tomaymycin biosynthetic pathway
|
| 17 | c2qsgX_ |
|
 |
7.5 |
22 |
PDB header:dna binding protein/dna
Chain: X: PDB Molecule:uv excision repair protein rad23;
PDBTitle: crystal structure of rad4-rad23 bound to a uv-damaged dna
|
| 18 | d2a7wa1 |
|
 |
7.2 |
14 |
Fold:all-alpha NTP pyrophosphatases
Superfamily:all-alpha NTP pyrophosphatases
Family:HisE-like (PRA-PH) |
| 19 | c2x4kB_ |
|
 |
6.8 |
5 |
PDB header:isomerase
Chain: B: PDB Molecule:4-oxalocrotonate tautomerase;
PDBTitle: crystal structure of sar1376, a putative 4-oxalocrotonate2 tautomerase from the methicillin-resistant staphylococcus3 aureus (mrsa)
|
| 20 | c2a7wF_ |
|
 |
6.7 |
14 |
PDB header:hydrolase
Chain: F: PDB Molecule:phosphoribosyl-atp pyrophosphatase;
PDBTitle: crystal structure of phosphoribosyl-atp pyrophosphatase2 from chromobacterium violaceum (atcc 12472). nesg target3 cvr7
|
| 21 | d1x3zb1 |
|
not modelled |
6.5 |
25 |
Fold:XPC-binding domain
Superfamily:XPC-binding domain
Family:XPC-binding domain |
| 22 | c2wb1J_ |
|
not modelled |
6.2 |
19 |
PDB header:transcription
Chain: J: PDB Molecule:dna-directed rna polymerase rpo13 subunit;
PDBTitle: the complete structure of the archaeal 13-subunit dna-2 directed rna polymerase
|
| 23 | c3mb2G_ |
|
not modelled |
6.1 |
10 |
PDB header:isomerase
Chain: G: PDB Molecule:4-oxalocrotonate tautomerase family enzyme - alpha subunit;
PDBTitle: kinetic and structural characterization of a heterohexamer 4-2 oxalocrotonate tautomerase from chloroflexus aurantiacus j-10-fl:3 implications for functional and structural diversity in the4 tautomerase superfamily
|
| 24 | d2fufa1 |
|
not modelled |
6.1 |
41 |
Fold:Origin of replication-binding domain, RBD-like
Superfamily:Origin of replication-binding domain, RBD-like
Family:The origin DNA-binding domain of SV40 T-antigen |
| 25 | c2p9xB_ |
|
not modelled |
6.1 |
50 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hypothetical protein ph0832;
PDBTitle: crystal structure of ph0832 from pyrococcus horikoshii ot3
|
| 26 | c2ormA_ |
|
not modelled |
5.9 |
10 |
PDB header:isomerase
Chain: A: PDB Molecule:probable tautomerase hp0924;
PDBTitle: crystal structure of the 4-oxalocrotonate tautomerase homologue dmpi2 from helicobacter pylori.
|
| 27 | c3n4dF_ |
|
not modelled |
5.7 |
5 |
PDB header:hydrolase
Chain: F: PDB Molecule:putative tautomerase;
PDBTitle: crystal structure of cg10062 inactivated by(r)-oxirane-2-carboxylate
|
| 28 | d1y6xa1 |
|
not modelled |
5.7 |
45 |
Fold:all-alpha NTP pyrophosphatases
Superfamily:all-alpha NTP pyrophosphatases
Family:HisE-like (PRA-PH) |
| 29 | d1mwwa_ |
|
not modelled |
5.2 |
10 |
Fold:Tautomerase/MIF
Superfamily:Tautomerase/MIF
Family:Hypothetical protein HI1388.1 |
| 30 | c3abfB_ |
|
not modelled |
5.2 |
15 |
PDB header:isomerase
Chain: B: PDB Molecule:4-oxalocrotonate tautomerase;
PDBTitle: crystal structure of a 4-oxalocrotonate tautomerase homologue2 (tthb242)
|

