Conservation of Structural Features
What follows is some work I did in Oxford with
Geoff Barton. You can read this
paper by clicking here.
In all figures shown below, pairs of proteins are shown which adopt similar
three-dimensional (3D) structures in the absense of any apparent sequence
similarity. All structure are shown side-by-side in a superimposed orientation.
Regions of the structure that are structurally equivalent are shown as arrows, ribbons
or coils; structurally unconserve regions are shown as C-alpha trace. The structures are
coloured from Blue-to-Red as the chain progresses from N- to C- terminus. Below eac
pair of structures is the correponding structurally-based sequence alignment.
Structurally equivalent regions are boxed. Those residues having equivalent values
for each of the three structural features discussed are shown in pink. More
details are given below.
Type B similarities refer to those pairs of protein 3D structures having structural and functional similarities (e.g. eukaryotic and viral aspartic proteinases; globins from root nodules and mammals; etc.). Type C similarities refer to those pairs of protein 3D structures having only 3D structural similarity (i.e. no sequence or functional similarity). Type B similarities are usually argued to be related by divergent evolution, wherease type C similarities are argued either way (depending on ones opinion). The figures show that structural conservation bears no relation to functional similarity; often proteins having no functional similarity have a degree of structural feature (e.g. accessibility, secondary structure, etc.) conservation comparable to, or even higher than, proteins having functional similarity.
See Russell, R.B. & Barton, G.J. (1994) J. Mol. Biol. , 244, 332-350.
Protein Core conservation
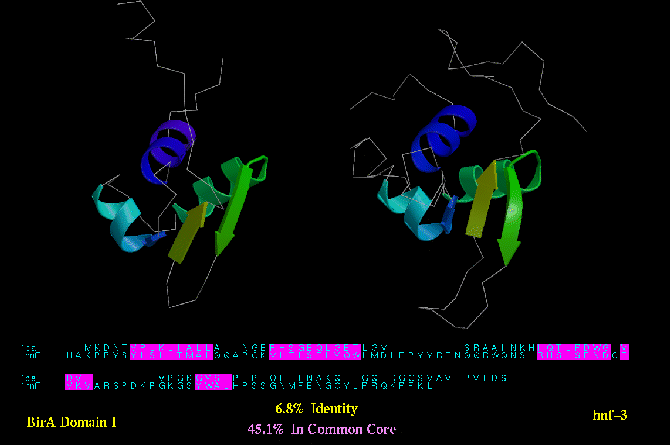
Type B similarity: Biotin operon protein (BirA; PDB code 1BIA) and HNF-3 (below) are both DNA binding domains with a
common helix-turn-helix motif. Only 45.1 % of their residues can be deemed
structurally equivalent.
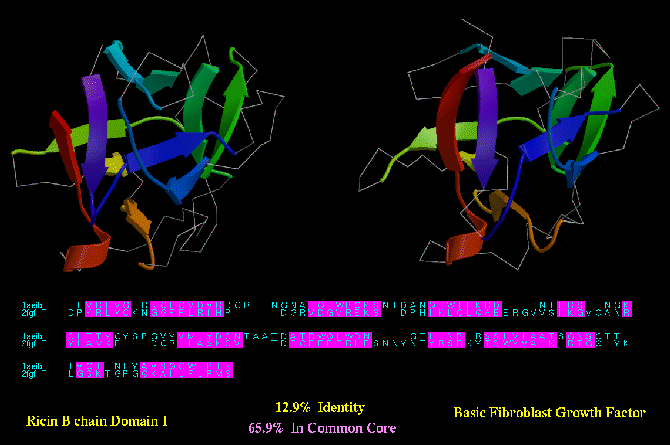
Type C similarity: Ricin (1AAI_A) and Basic Fibroblast Growth factor (2FGF; below) have a much higher degree of structurally
equivlent residues (almost 66 %). However, unlike the BirA/HNF-3 example, these
two proteins have no apparent functional similarity.
Accessibility conservation
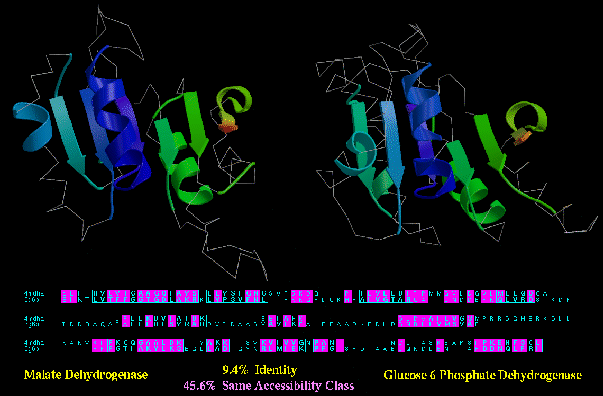
Type B similarity: malate and glucose-6-phosphate dehydrogenase (4MDH_A and 2G6P; below) have obvious
functional similarity: they both bind NAD or NADP, they catalyse similar reactions,
and they even have a few regions of local sequence similarity (e.g. the GXGXXG/A loop near
the N-termini). In spite of this, they only have 45.6% of their residues in the same
accessibility class (e.g. buried, half-bured, or exposed).
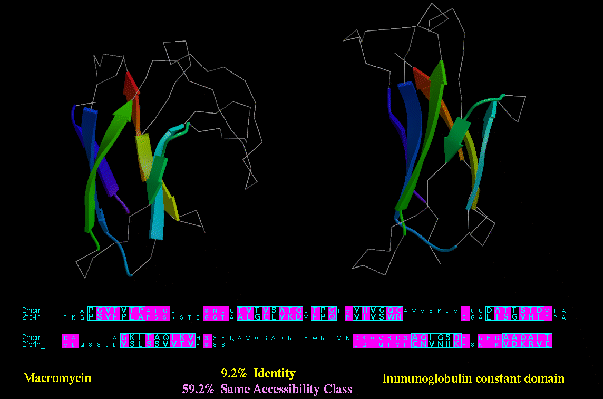
Type C similarity: Macromycin (2MCM) and an Immunoglobulin constant domain (2FBJ_L; below), on the other hand, have a much higher degree of residues in the same accessibility class (59.2%), despite no apparent functional or
sequence similarity.
Secondary structure conservation
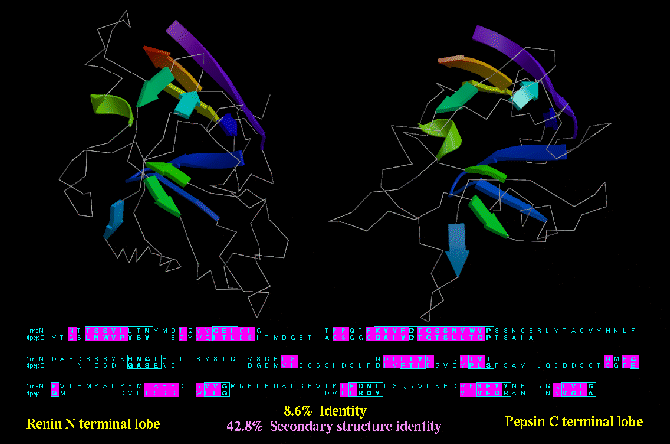
Type B similarity: the N- and C- terminal lobes of aspartyl proteinases are very likely to have
diverged from a common ancestor. The N- terminal lobe of Renin (1RNE) and the C-terminal lob ov
Pepsin (2PEP) are shown below. Not only are they similar structures, but they both contribute
the same residue (i.e. an aspartic acid) to a common active site. Moreover, the existence of
monomeric aspartyl proteinases in viruses (i.e. which dimerise rather than consist of
a two domain protein) reinforces the relationship between N- and C- terminal lobes. Despite
this relationship, only 42.8 % of the residues are of the same secondary structure type.
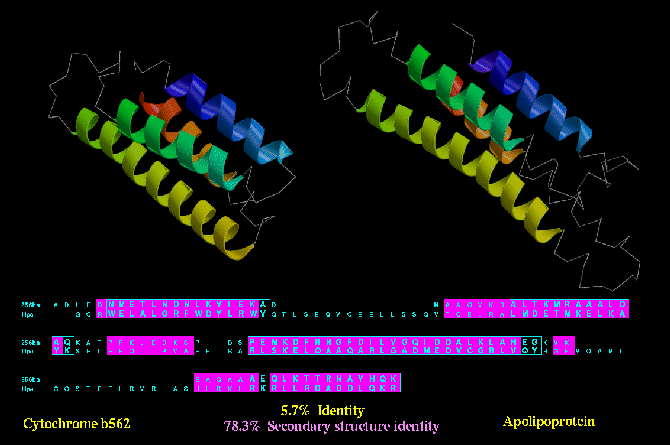
Type C similarity: Cytochrome b562 (256B_A) and Apolipoprotein (1LPE) are both four helix
bundles, and despite a lack of sequence or functional similarity, these two protein show a
higher degree of secondary structure identity (78.3 %) than the type B similarity shown
above.