Sternberg Group Binding Site Prediction in CASP8
See Wass, M.N. and Sternberg, M.J. (2009) Prediction of ligand binding sites using homologous structures and conservation at CASP8. Proteins, 77 Suppl 9:147-51. PubMed
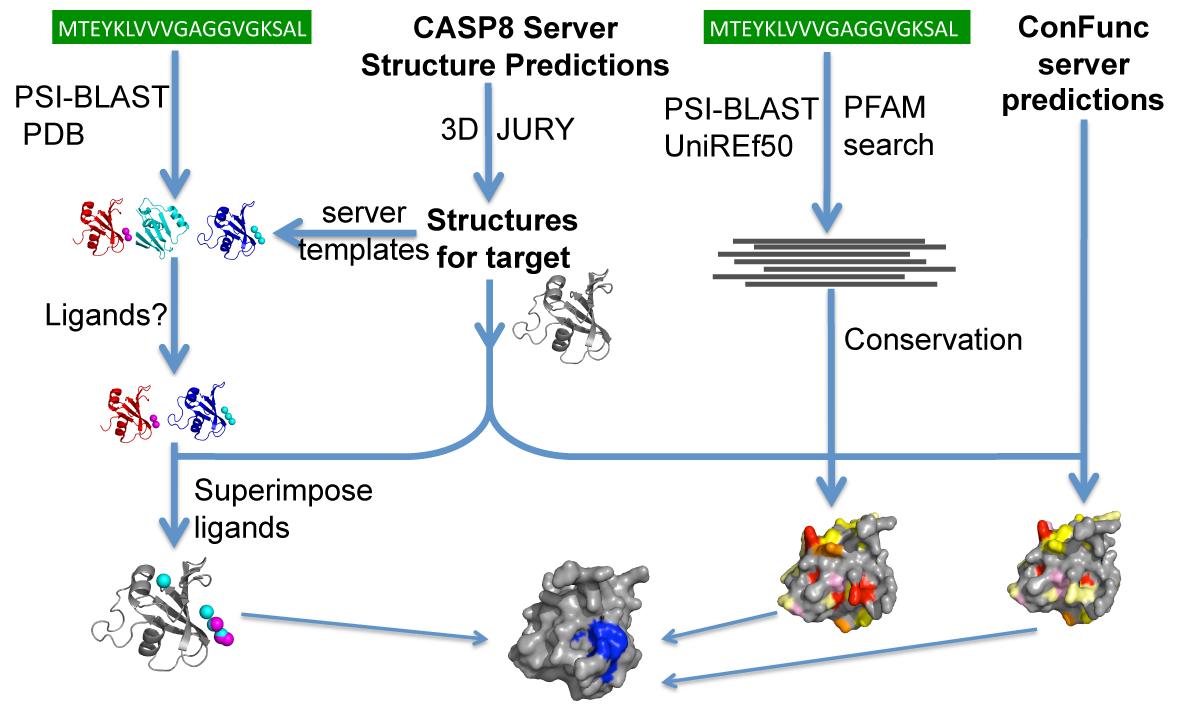
The diagram below outlines our approach for ligand binding site prediction at CASP8. We used the server structure predictions as models of the target and superimposed the ligands from homologous structures on to the model. In addition we calculated conservation from ConFunc and from Pfam hits to the target. We have developed a server, 3DLigandSite which automates our method.

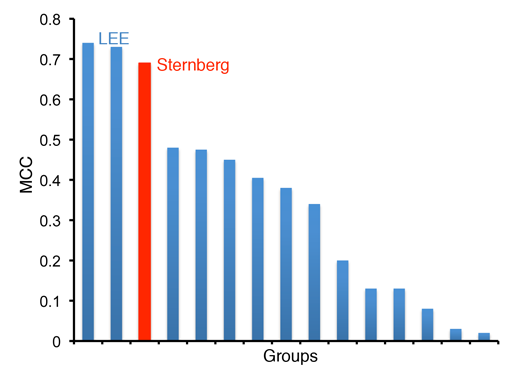
Overall we were one of the top performing groups, with only the LEE group obtaining better results (although these differences were not significantly different (See assessment paper - Lopez et al., 2009 - Pubmed). The graph below shows the MCC for each group, the Sternberg group is shown in Red. This figure is adapted from Lopez et al.,
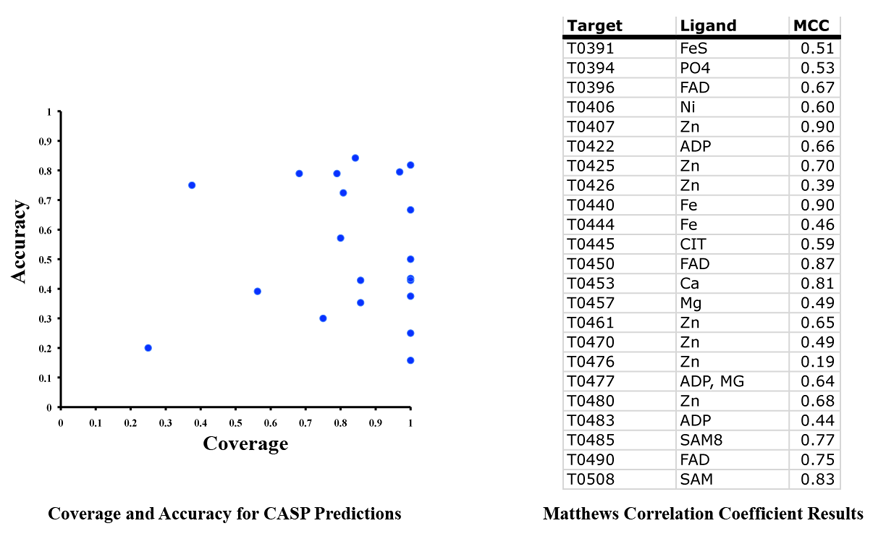
We have analysed our results using the Matthews Correlation Coefficient and measures of Coverage (% of binding site that has been predicted) and Accuracy (% of all residues predicted that are correct). Overall we obtained an MCC of 0.63, 82% coverage and 56% accuracy. The results for each individual prediction are shown below.
Visualisation of our predictions
Some example of our predictions and their agreement with the binding site in the solved structure are shown below. The ligand is coloured magenta, True positive residues blue, Fale positive residues red and false negative residues yellow.
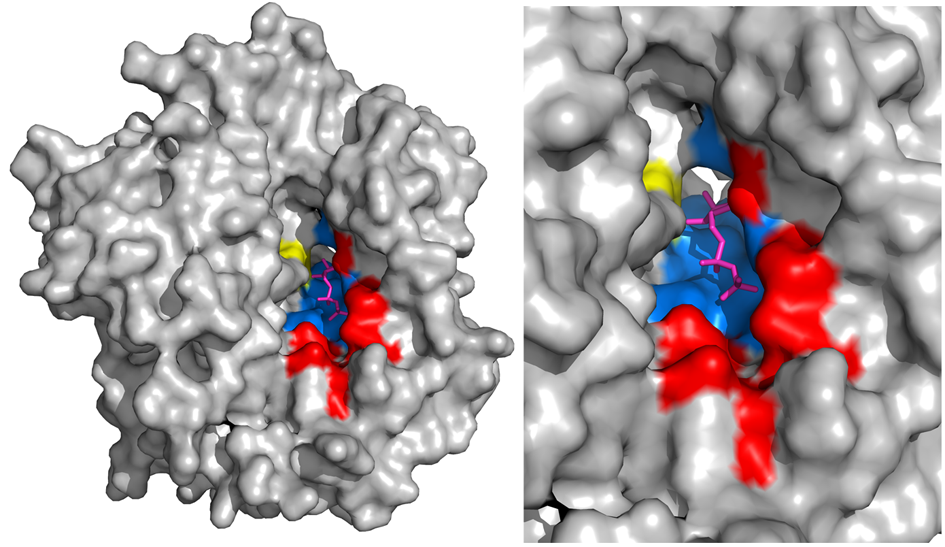
T0422 - AAA+ ATPase domain from Human Fidgetin-like protein (pdb code 3d8b). Ligand bound is ADP.
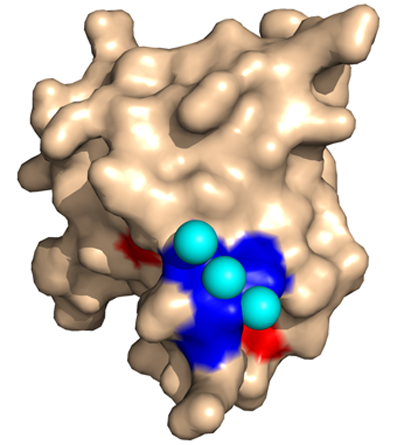
T0453 - 3 Calcium Ions (cyan) bound to target.