Robert B. Russell & Michael J. E. Sternberg (1997) Two new examples of protein structural similarities within the structure-function twilight zone, Protein Eng., 4, 333-338.
Two three-dimensional protein structural similarities accompanied by slight similarities in function are reported to highlight the present difficulties in discerning the relationship between structure and function. The similarity between the structures of uteroglobin and the CAP domain of haloalkane dehalogenase reveals a common four helix hydrophobic ligand binding motif. The similarity between the beta barrel domains of glutaminyl tRNA synthetase and domain 1 of glutamine synthetase is accompanied by some similarity in the location of nucleotide binding sites and suggests a possible ancient domain common to these two proteins involved in binding of glutaminyl moieties. The problems raised by these two similarities in the structure-function "twilight zone" are discussed.
Please see the paper for more details. Here are some colour pictures and further data (e.g. coordinates) illustrating the similarities discussed.

MSF or AMPS block or PostScript version of the alignment above.
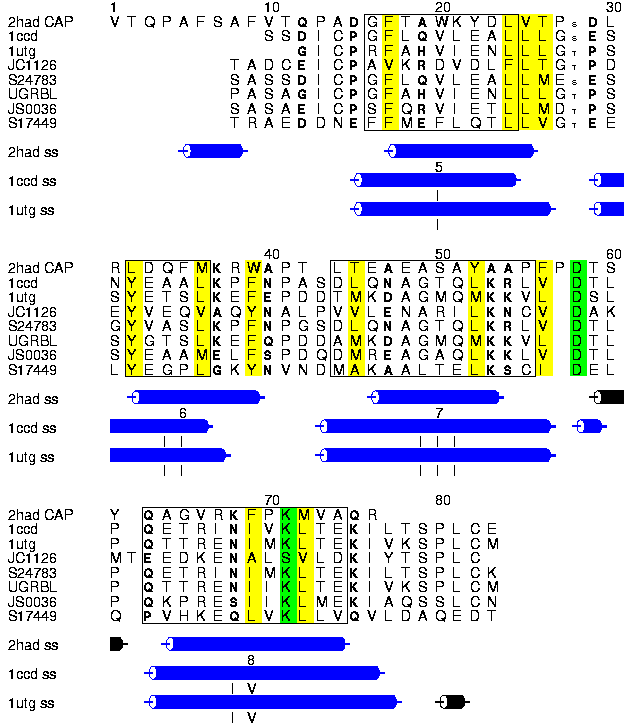
Alscript figure showing the structure-based alignment of the HAD CAP domain (2HAD residues 156-229), Rat Uteroglobin (1UTG) and its human homologue of known 3D structure Clara Cell protein (1CCD) together with an alignment of uteroglobin homologues (JC1126, S24783, UGRBL, JS0036, S17449). No haloalkane homologues could be found in sequence databases. Residues showing a conservation of hydrophobic, polar or small character are shown in yellow, bold or small font respectively. Positions were defined as conserved with respect to these three characters if these residues were observed in all positions with the allowed exception of one UTG homologue. Structurally equivalent regions are boxed. Residues known to contact substrates in UTG and HAD proteins are shown in inverse text. The locations of alpha and 3_10 helices within the known structures are shown below the alignment, and alpha helices numbered I-IV in UTG and 5-8 in HAD. The differences in helix orientation and length suggest an alternative structural alignment that would shift the first equivalenced helix in HAD 4 residues in an N-terminal direction relative to UTG.
The alignment and superimposition of the structures was performed by STAMP. Alignment of the uteroglobin sequence was done using AMPS.
Colour picture of the two domains + heteroatoms:
PDB format superimpositions:
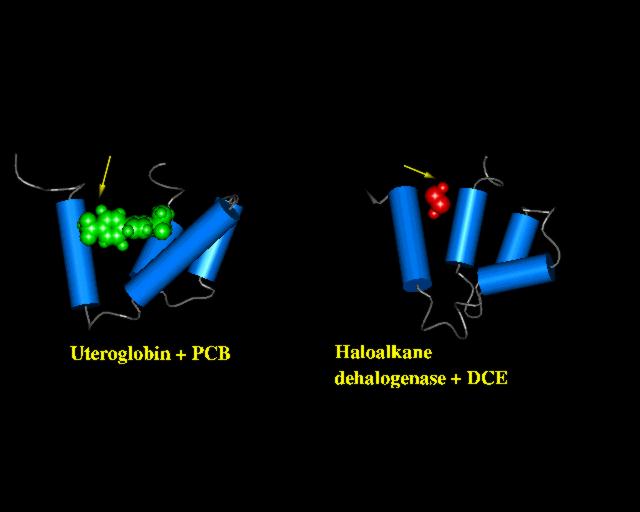
The PREPI figure above shows the two domains in a similar orientation. The arrows indicate the location of bound polychlorinated biphenyl (uteroglobin, green above) and dichloroethane (haloalkane dehalogenase, red above).
MSF or AMPS block or PostScript version of the alignment above.
In the text we discuss (but do not show) a larger alignment including GTRS and GS homologues, this alignment can be found here in MSF or AMPS block or PostScript formats.
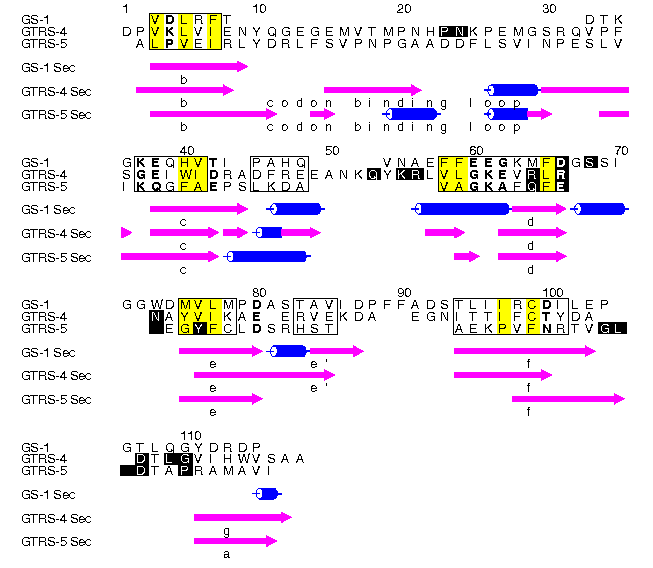
Alscript figure showing the structure-based alignment of the N-terminal domain of glutamine synthetase (1LGR residues 17-105) and domains 4 & 5 of glutaminyl tRNA synthetase (1GTR chain A; domain 4 residues 347-464; domain 5 residues 464-547, 339-347). Beta strands are shown as mangenta arrows; helices as blue cylinders. Secondary structures are labelled according to definitions found in the paper. Residues showing a conservation of hydrophobic, polar or small character are shown yellow, bold or small font respectively. Positions were defined as conserved with respect to these three characters if these residues were observed in all positions with one allowed exception in all homologues to GS or GTRS found. Positions shown in inverse text are show in contact with bound RNA or heteroatoms.
The alignment and superimposition of the structures was performed by STAMP. Alignment of the uteroglobin sequence was done using AMPS.
Colour picture of the three domains + heteroatoms:
PDB format superimpositions:
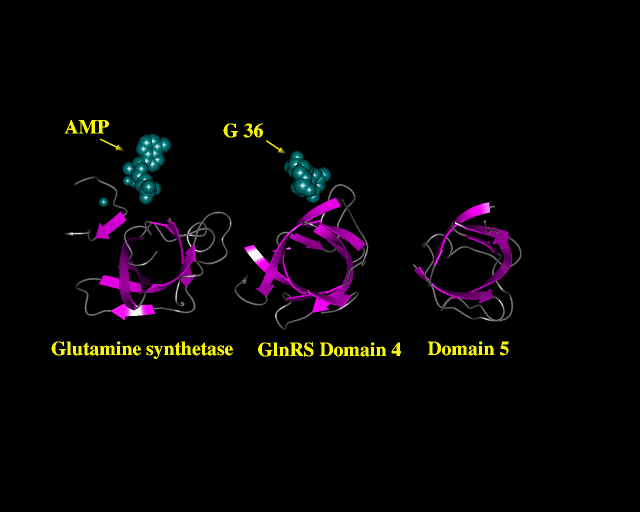
The PREPI figure above shows the three domains in a similar orientation. Spacefill shows the location of AMP and two metal ions in GS and a base of tRNA (G36) in an equivalent position in domain 4 of GTRS.