This page complements R.B. Russell & M.J.E. Sternberg, Protein Eng.,9, 107-111, 1996.
During an all-against-all comparison of a database of protein domains, we found a significant similarity between a domain in catalase and the structures of calycins, a group of proteins having no sequence similarity, but which adopt a common up-down-up-down, anti-parallel beta barrel structure.
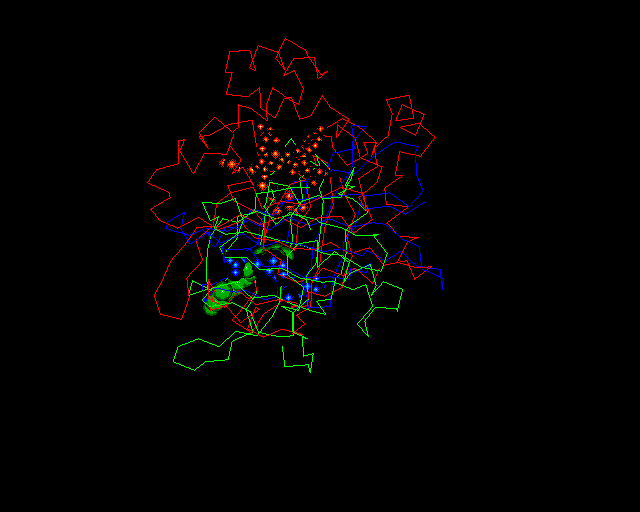
If you would like to a PDB file containing catalase superimposed onto retinol binding protein (RBP, a lipocalin) and avidin, complete with heteroatoms, etc., click here. The superimposition looks like:

Known members of the calycin family (including avidin and RBP) binding molecules in a similar pocket. Lipocalins and fatty acid binding proteins bind to small hydrophobic molecules, and avidins bind biotin with a high affinity. Catalase appears to have a similar, though partially covered pocket. The size of the pocket is shown below (middle) beside biotin (left) and salicylic acid (right), which is known to inhibit some catalases.